What we work on
In our research, we work with two model organisms: the threespine stickleback (a freshwater fish), and flies in the genus Drosophila, particularly D. melanogaster and D. pseudoobscura.
We employ a wide variety of research methods in the course of our research, including genomics (molecular and computational), field work, computer simulations, and experiments.
Current Projects
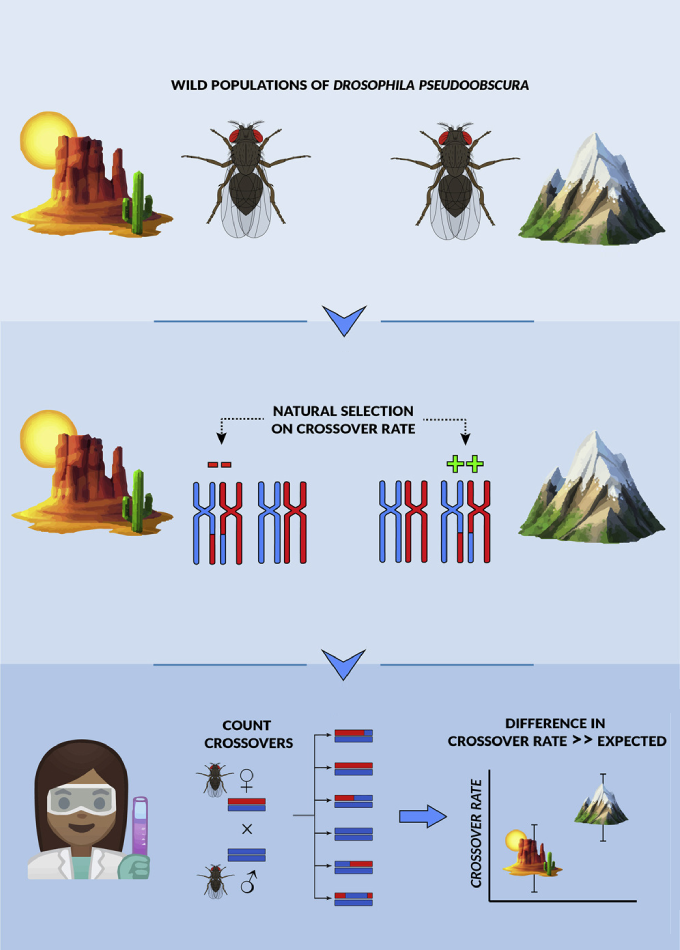
The evolution of recombination rate
Recombination rate, genome-wide or along a chromosome, is often treated as a static "species level" trait. However, recent work (including our own) has shown that natural populations vary considerably in recombination
rate,
and this variation
has a genetic basis. In other words, recombination rate is a heritable phenotype that evolves, and can be studied with the toolkit of evolutionary ecology.
We have several active projects aimed at understanding the evolution of recombination rate:
- Studying the genetic basis of recombination rate variation in natural populations
- Exploring the role natural selection plays in driving differences in recombination rate among populations
- Experimental studies of the evolution of recombination modifiers
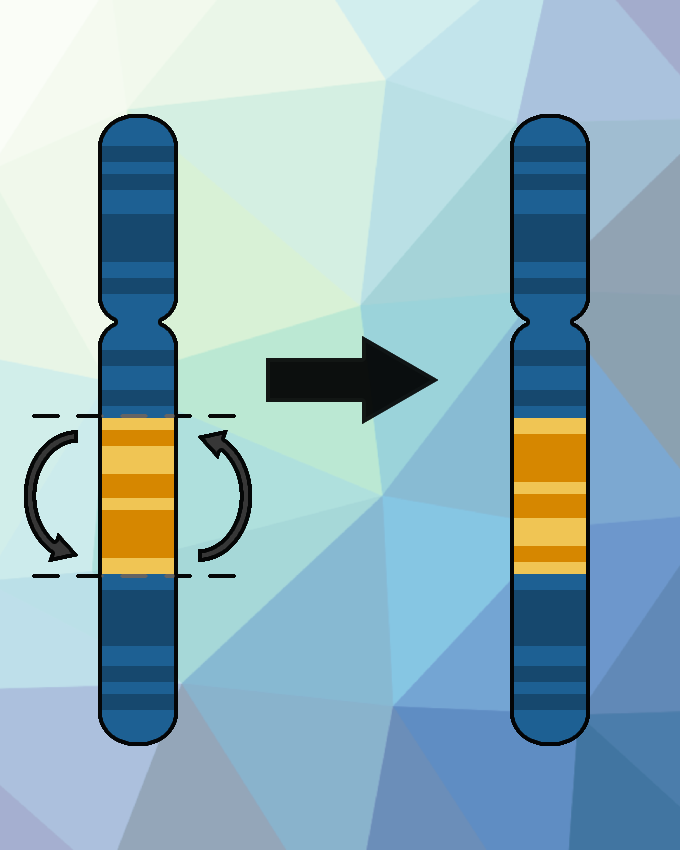
Chromosomal Inversions
Chromosomal inversions are a class of structural variant in which a segment of chromosome is flipped/reversed in orientation relative to the reference sequence. Studies over the last 100 years have found that inversions
often
underlie local adaptation and reproductive isolation and are widely believed to evolve via natural selection.
Yet, in spite of a long history of study, the specific role chromosomal inversions play
during adaptation remains unclear. Our current efforts to clarify the role of inversions in adaptation include:
- Creating synthetic chromosomal inversions using modern transgenic approaches.
- Testing hypotheses about inversion evolution using experimental evolution,
- Field studies of the dynamics of inversions in natural populations of D. pseudoobscura from California

Accelerating breeding with recombination modifiers
Adaptation to new environments can be limited by the availability of optimal haplotypes — haplotypes that contain all the "Good" alleles that
increase traits of interest, and none of the "Bad" alleles that result in unwanted changes. A key parameter shaping haplotypic diversity is recombination rate.
We are working to explore how recombination rate can be optimized to accelerate artificial selection in agricultural contexts.
- How recombination rate could be optimized to accelerate various artificial selection regimes
- New methods for modifying recombination rate in breeding populations
- Experimental studies of optimized recombination in laboratory populations